Vol. 7, Issue 1 (2019)
Genome wide identification of genic and non-genic Microsatellites in Nilaparvata lugens Stål
Author(s): Rakesh Bhowmick, Soham Choudhury, ARNS Subbanna, Suman Roy and Laxmi Sharma
Abstract: The brown plant hopper (BPH), Nilaparvata lugens Stål (Homopterous: Delphacidae) is one of the lethal pests in rice. In the present study, genome wide distributed Simple Sequence Repeats (SSR) were identified and analysed. The screening of 1.2 Gb of BPH genome resulted in identification of 271785 SSRs. Out of these, 3231 SSRs were from genic region. Microsatellite distribution in genome was found to be 238.2 SSRs per Mb. Altogether the microsatellite motifs contribute only 7.45Mb of BPH genome. Trinucleotide repeats were most abundant representing 50.8% of total genomic microsatellites. Longer repeat motifs were found to be least abundant in the genome representing 1.1% and 0.5% for pentanucleotide and hexanucleotide, respectively. SSR markers identified from this study can be useful for diversity analysis and genetic map development of BPH.
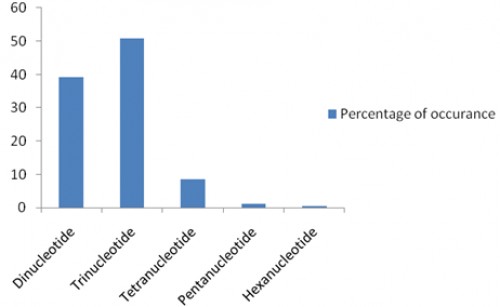
Fig. 1: Distribution with respect to the motif repeats of di to hexa-nucleotide microsatellites
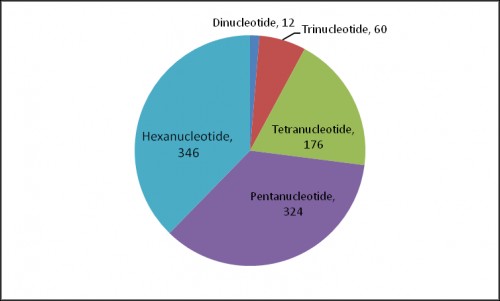
Fig. 2: Distribution of microsatellite motif length variants. Pie diagram represents motif length of 918 different microsatellite variants
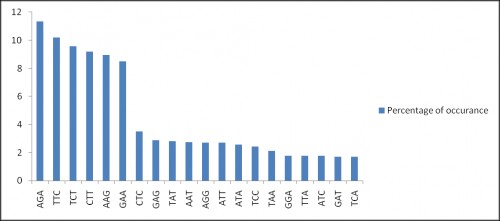
Fig. 3: Distribution of trinucleotide repeat motifs in genome. Y axis represents percentage of occurrence in genome
Pages: 194-196 | 739 Views 56 Downloads
download (4375KB)
How to cite this article:
Rakesh Bhowmick, Soham Choudhury, ARNS Subbanna, Suman Roy, Laxmi Sharma. Genome wide identification of genic and non-genic Microsatellites in Nilaparvata lugens Stål. Int J Chem Stud 2019;7(1):194-196.